Welcome to Ion Mobility – Mass Spectrometry (IM-MS) website from the Zhu Lab, Chinese Academy of Sciences, Shanghai.
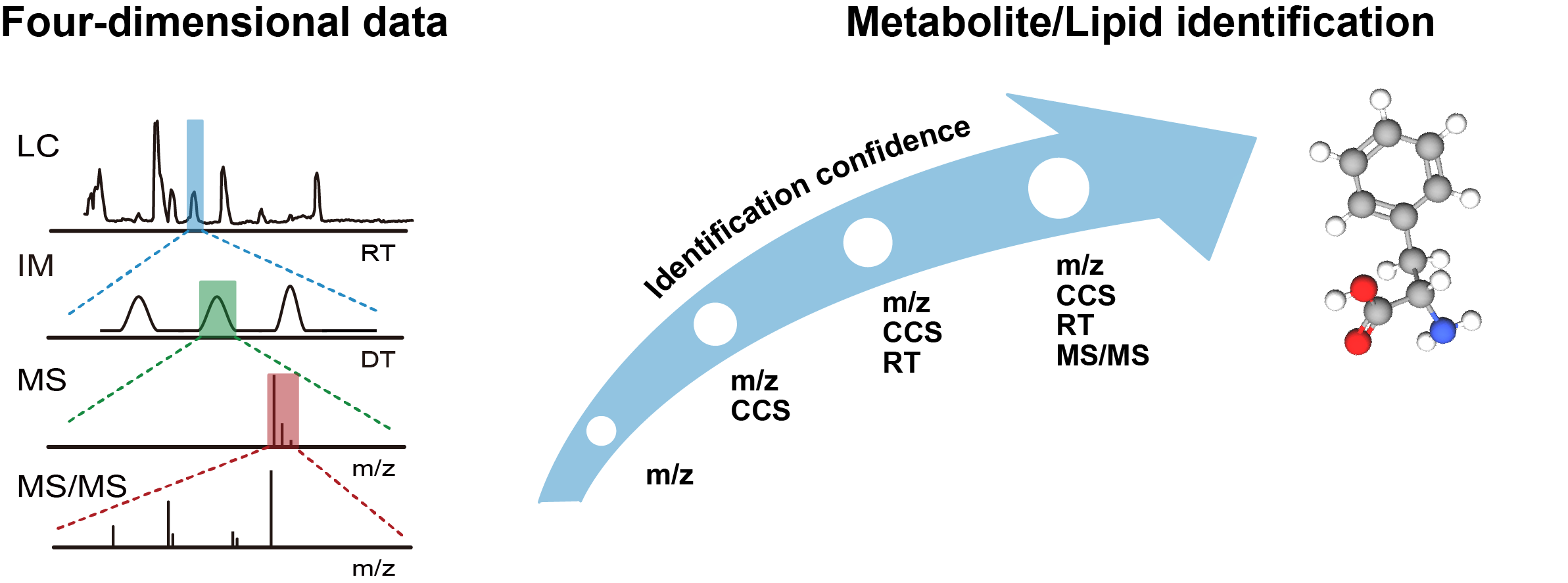
IM-MS has recently been introduced to analyze the complex biological samples towards metabolomics and lipidomics applications. IM technology utilizes the collisions between ions and inert buffer gas under an electric field to rapidly separate ions based on their structural and conformational differences. It provides the orthogonal separation to effectively reduce chemical noise, improve the signal-to-noise, and enhance peak capacity. It also enables to distinguish the isomeric metabolites and lipids that commonly exist in the biological samples. The use of IM-MS for metabolomics and lipidomics has facilitated the separation and the identification of metabolites and lipids in complex biological samples. The collision cross-section (CCS) value derived from IM-MS is a valuable physiochemical property for the unambiguous identification of metabolites and lipids.
Software tools & Database
In this website, we provide several important software tools and databases developed in Zhu Lab to support the application of IM-MS in metabolomics and lipidomics.

MetCCS is a free webserver to predict the CCS values of METBOLITES. MetCCS also provides a database with predicted CCS values for 35,203 metabolites from HMDB. One can easily search and match these CCS values in the webserver.
Go to visit
LipidCCS is a free webserver to predict the CCS values of LIPIDS. The prediction can be made using the SMILES structure as an input. LipidCCS also provides a database with predicted CCS values of 15,646 lipids from LIPID MAPS.
Go to visit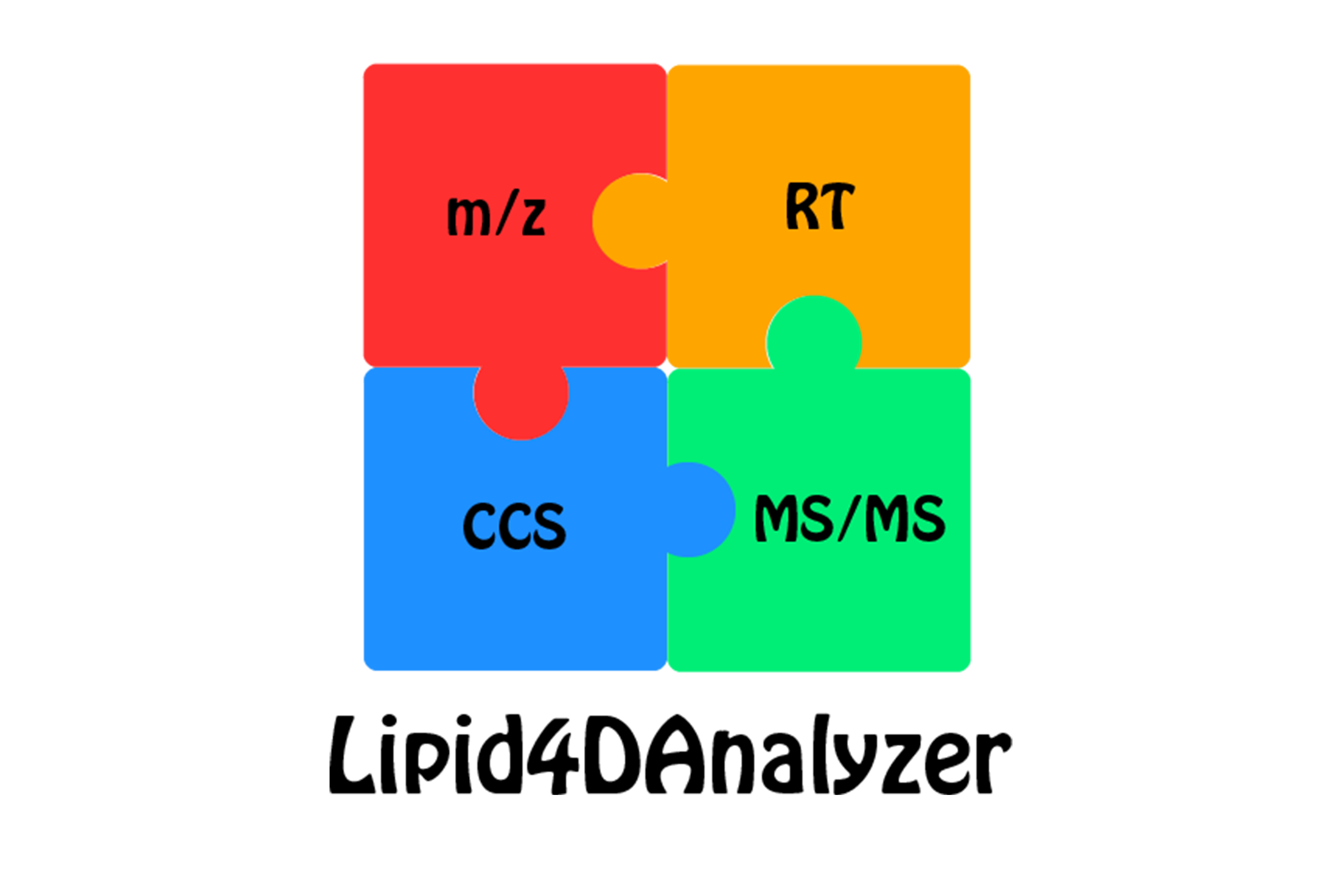
Lipid4DAnalyzer is an upgraded version of LipidIMMS Analyzer (since August. 10, 2021) to support lipid identifications in MS based lipidomics. It intergrates milti-dimensional information including m/z, RT, CCS and MS/MS spectra for the ambiguous identification of lipids.
Go to visit
AllCCS is a powerful platform to support various applications in IM-MS, containing three major parts: 1) Unified CCS database (~1,700,000 small molecules), 2) CCS prediction, and 3) Match and rank function for small molecule annotation.
Go to visitPublications
AllCCS:
1. Z. Zhou, M. Luo, X. Chen, Y. Yin, X. Xiong, R. Wang, and Z.-J. Zhu* (Corresponding author), Ion Mobility Collision Cross-Section Atlas for Known and Unknown Metabolite Annotation in Untargeted Metabolomics, Nature Communication, In pressing.
Note: AllCCS database, CCS prediction and combined strategy for identifying knowns and unknowns
2. H. Tsugawa*, K. Ikeda, M. Takahashi, A. Satoh, Y. Mori, H. Uchino, N. Okahashi, Y. Yamada, I. Tada, P. Bonini, Y. Higashi, Y. Okazaki, Z. Zhou, Z.-J. Zhu, J. Koelmel, T. Cajka, O. Fiehn, K. Saito, M. Arita, and M. Arita* (Corresponding author), A lipidome atlas in MS-DIAL 4, Nature Biotechnology, 2020, 1–5.
Note: MS-DIAL 4, IM-MS data process and multi-dimensional identification
IM-MS based Metabolomics:
3. Z. Zhou, X. Shen, J. Tu, and Z.-J. Zhu* (Corresponding author), Large-Scale Prediction of Collision Cross-Section Values for Metabolites in Ion Mobility - Mass Spectrometry, Analytical Chemistry, 2016, 88, 11084-11091.
Note: MetCCS, machine-learning based CCS prediction
4. Z. Zhou, X. Xiong, and Z.-J. Zhu* (Corresponding author), MetCCS Predictor: A Web Server for Predicting Collision Cross-Section Values of Metabolites in Ion Mobility-Mass Spectrometry based Metabolomics, Bioinformatics, 2017, 33, 2235-2237.
Note: MetCCS predictor, webserver for CCS prediction
5. Z. Zhou†, J. Tu†, and Z.-J. Zhu* (†, Co-first authors; *, Corresponding author), Advancing the Large-Scale CCS Database for Metabolomics and Lipidomics at the Machine-Learning Era, Current Opinion in Chemical Biology, 2018, 42, 34-41.
Note: Review on approach for generating CCS database and advantage of using machine-learning based CCS prediction
6. M. Luo, Z. Zhou, and Z.-J. Zhu* (Corresponding author), The Application of Ion Mobility-Mass Spectrometry in Untargeted Metabolomics: from Separation to Identification, Journal of Analysis and Testing, 2020, 4, 163–174.
Note: Review on application of IM-MS in untargeted metabolomics
IM-MS based Lipidomics:
7. Z. Zhou, J. Tu, X. Xiong, X. Shen, and Z.-J. Zhu*(Corresponding author), LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision to Support Ion Mobility-Mass Spectrometry based Lipidomics, Analytical Chemistry, 2017, 89, 9559–9566.
Note: LipidCCS, machine-learning based CCS prediction for lipids
8. J. Tu, Y. Yin, M. Xu, R. Wang and Z.-J. Zhu* (Corresponding Author), Absolute quantitative lipidomics reveals lipidome-wide alterations in aging brain, Metabolomics, 2018, 14, 5.
Note: LipidAnalyzer, LC-MS based absolute quantitative lipidomics
9. Z. Zhou, X. Shen, X. Chen, J. Tu, X. Xiong, and Z.-J. Zhu* (Corresponding Author), LipidIMMS Analyzer: Integrating Multi-dimensional Information to Support Lipid Identification in Ion Mobility–Mass Spectrometry based Lipidomics, Bioinformatics, 2019, 35,698-700.
Note: LipidIMMS Analyzer, multi-dimensional database and identification tool for lipids on IM-MS
10. J. Tu†, Z. Zhou†,T. Li†, and Z.-J. Zhu* (Corresponding Author) (†, Co-first authors; *, Corresponding author), The Emerging Role of Ion Mobility-Mass Spectrometry in Lipidomics to Facilitate Lipid Separation and Identification, Trends in Analytical Chemistry, 2019, 116, 332-339.
Note: Review on application of IM-MS in untargeted lipidomics
11. X. Chen, Z. Zhou, and Z.-J. Zhu* (Corresponding author), The Use of LipidIMMS Analyzer for Lipid Identification in Ion Mobility-Mass Spectrometry-Based Untargeted Lipidomics, Methods in Molecular Biology, 2020, 2084, 269-282.
Note: LipidIMMS Analyzer protocol
12. X. Chen†, Y. Yin†, Z. Zhou, T. Li, and Z.-J. Zhu* (†, Co-first authors; *, Corresponding author), Development of A Combined Strategy for Accurate Lipid Structural Identification and Quantification in Ion-Mobility Mass Spectrometry based Untargeted Lipidomics, Analytica Chimica Acta, 2020, In Revision.
Note: Lipid4DAnalyzer, combined multi-dimensional match and rule decision for lipidomics identification on IM-MS